What are we doing?
Project collaborators Dr Susana Sauret-Gueto and Dr Eftychios Frangedakis, from the University of Cambridge.
We live in an era in which we can thoroughly investigate all the genetic material that makes up an organism, at the level of the whole genome. Understanding gene regulation, the set of processes that control the decoding of DNA, is an essential part of modern synthetic biology. Although the expression of a gene can be regulated at different levels, transcription is one of the most important steps in this complex multistage process. Transcription is the process of creating messenger sequences, known as RNA, which allow the translation machinery of the cell to build proteins according to the DNA instructions. The efficiency of transcription determines of how much of these messenger RNAs are produced.
Transcription is triggered by a collection of functional proteins known as transcription factors (TFs), which bind onto the DNA sequence in front of the part of the gene that encodes a protein (the coding sequence). This section of DNA is called the promoter. Previous research has demonstrated that the strength of a binding event between a TF and the part of a DNA sequence in the promoter that it can ‘recognise’, is pivotal in affecting the transcription of the associated gene. The strength of this binding event between a TF and its binding site (TFBS) is referred to as binding affinity. Our OpenPlant funded project, based at the Earlham Institute and the University of Cambridge, is focused of finding out how much variation in binding affinity exists in nature, and subsequently creating synthetic promoters with varying binding affinities. We aim to develop a new method to test how variation in TFBS sequence might affect binding affinity. With the knowledge gained, we can then build promoter sequences that activate transcription at the level we design, in the place in a plant we want.
Methods of assessing the binding affinity of TFs and DNA sequences already exist, but have limited scope for asking questions such as ours, either due to low throughput or high cost. One such method, which has been established for decades, is the Electrophoretic Mobility Shift Assay (EMSA). EMSA is based on visualising the travel of the bound TF-DNA complex through a jelly like matrix, or gel. Because each sequence and TF have to be made, and then individually ‘run’ through the EMSA gel, this method is difficult to scale up.
Figure 1: Comparing current protein-DNA binding assays.
Another option for testing the binding affinity of TFBS is based on a high-density DNA chip, also known as DNA microarray. This is essentially a glass slide with hundreds of thousands of short DNA sequences attached to the surface. The TF of interest can be synthesised in the lab and then hybridised with the chip. Hybridisation is just letting the binding between TF and TFBS occur as is would in the cell and, following this, the amount of TF bound on to each DNA sequence can be detected. However, the process of creating and reading such a chip is expensive and needs specific devices (Figure 1).
The aim of our work is to design and test a methodology which overcomes the shortcomings of available methods for testing TF binding affinity. Our goal is to provide a medium-throughput test of binding affinity, and we are designing our methodology to be easily replicable using affordable, readily available components. Approaching the problem from both synthetic and evolutionary backgrounds, we want to be able to test a range of binding sites for affinity, with enough replication to validate hypotheses. To do this we have developed the transcription factor relative affinity measurement pipeline (TRAMP).
Where did the ideas come from?
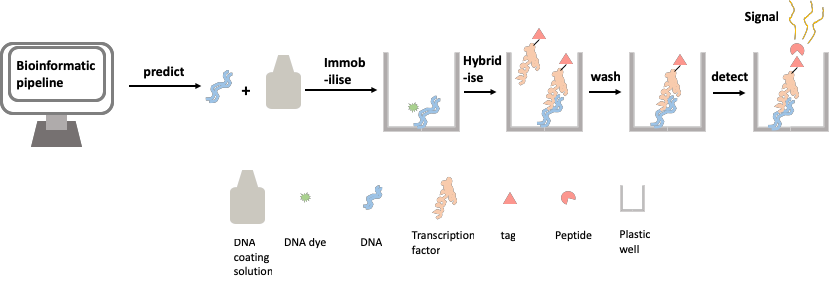
Inspired by the design of the DNA chip-based methods described above, we set out to design a method for assessing TF-DNA binding with as high a throughput as possible, whilst avoiding the high cost and specific requirements of the chip-based assay. We use a simple and cheap commercially available product to immobilise DNA onto the widely available lab workhorse; the 96-well plate. We have also used a recently developed method to tag our TF protein with a small peptide that, when bound to another peptide, gives off a signal. This signal is detectable in a plate reader, a piece of equipment widely used in modern labs. This new method allows us to easily quantify the amount of TF binding at a given TFBS, by measuring the brightness of the glow given off by the cumulative amount of the signal. These two components have allowed us to create a biochemical method of assaying protein-DNA binding affinity (Figure 2).
Figure 2: Assaying protein-DNA binding affinity using the transcription factor relative affinity measurement pipeline (TRAMP)
To maximise the efficiency of selecting TFBS to test in the assay we are designing, we have developed a novel computational tool. This allows us to leverage naturally occurring genetic variation in TFBS sequences gathered from publicly available population wide datasets. Instead of assaying thousands of random DNA sequences before finding a desirable one, we use several analytical methods to categorise natural variation along several parameters. We have been able to use a computational approximation of the binding event itself to score potential affinity. We also model the shape of the DNA double helix where the TFBS lies, allowing us to investigate the role of this physical property of DNA that has been suggested to be important in the efficiency of binding events.
These methods allow us to rapidly generate a set of suggested TFBS that should exhibit a range of binding affinities. This set of TFBS can be passed into the plate-based assay to be tested, and the findings used to test and further develop our understanding of binding affinity.
Where can this assay be applied?
We think that our work will be of interest to a wide range of biologists looking to understand the role of TF binding affinity in gene regulation. We hope by using TRAMP, we can find DNA sequences that exhibit different binding affinities to their corresponding TFs. Our aim is to be able to then replace the native TFBS in a promoter with the sequences we discover. We will use this approach to validate our findings, by linking the predictions and lab assays we have conducted back to transcription, the biological function we are interested in understanding.
It is our hope that our synthetic promoters will allow us to alter the activity of the promoter, meaning that when transcription occurs, the target gene will generate a different amount of messenger-RNA. If we can do this, we think our work will help scientists who want to use very specific gene expression patterns as part of synthetic biology strategies to synthesise valuable compounds like medicines. Our pipeline could also be useful for testing the effect of variation in TFBS between individuals, populations, or species.
By Dr Yaomin Cai and Dr Will Nash, Postdoctoral researchers at the Earlham Institute.