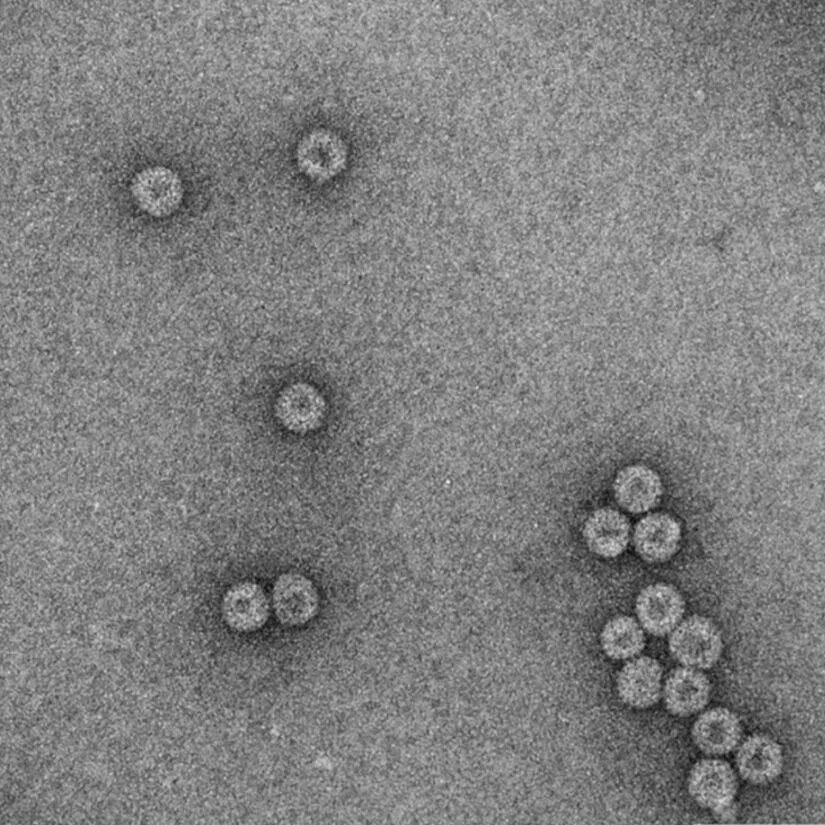
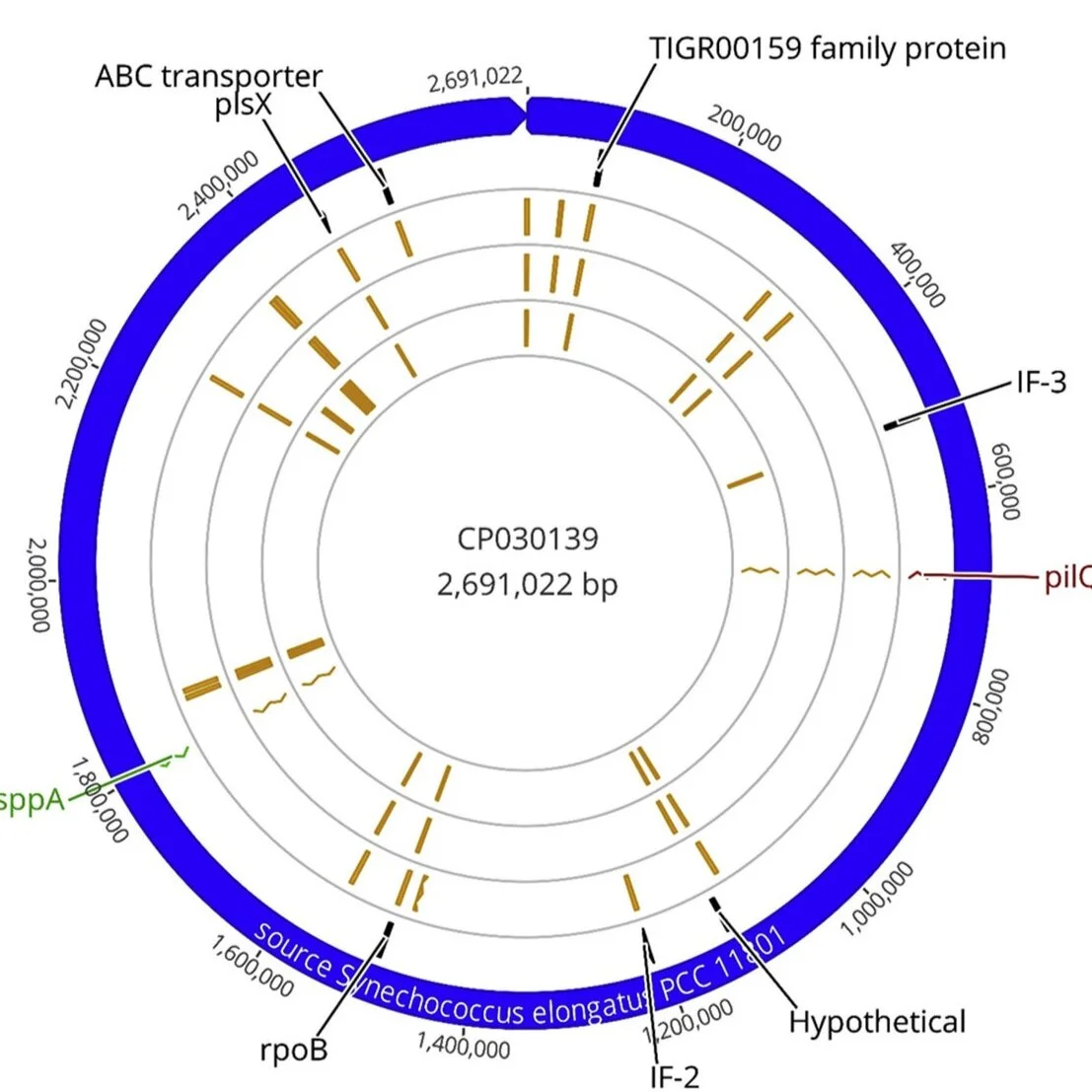
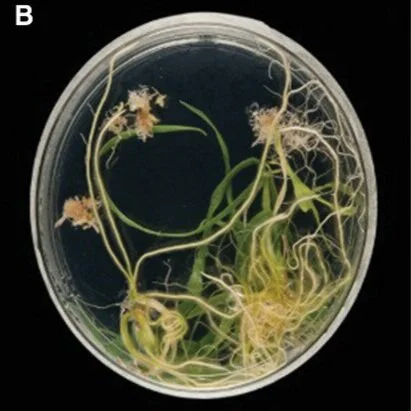
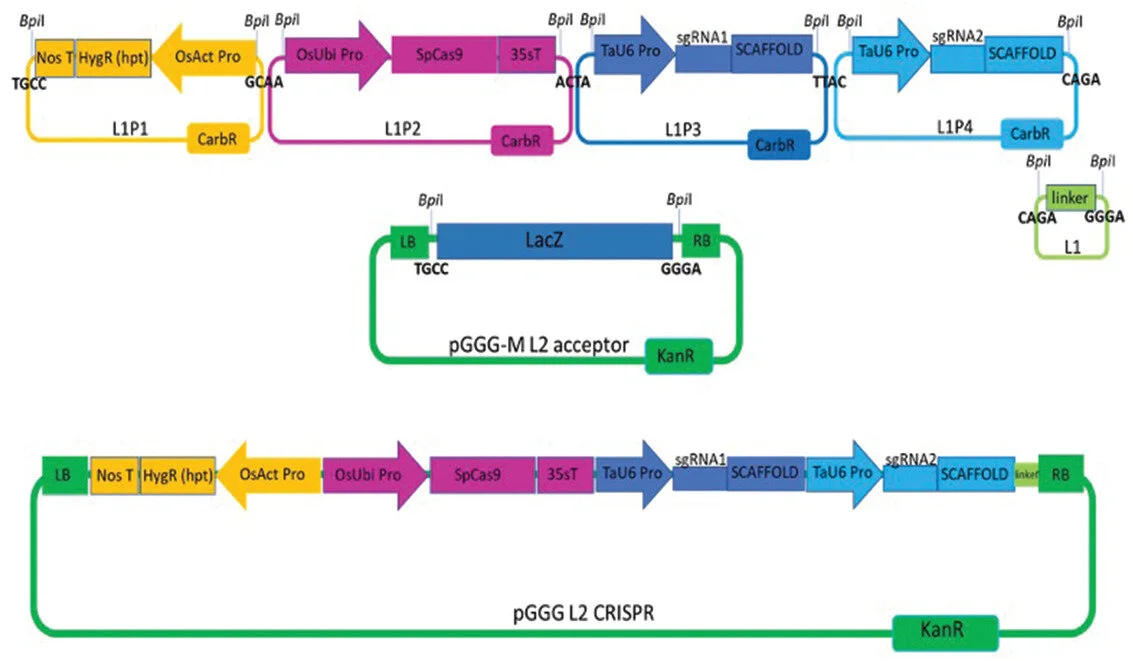
The Churchill Trust is proud to announce that a new Churchill Fellowship will be offered from 2022 to honour the memory and legacy of the late Saskia Beer, made possible through the generous sponsorship of Colin and Maggie Beer.
Churchill Fellowships are an internationally recognised award, providing access to expertise from around the world. Applicants are empowered to design their own projects, enabling them to explore international best practice and innovation that can be applied in Australia. There is a high level of visibility and credibility associated with becoming a Churchill Fellow, as well as a responsibility to share the knowledge and skills gained overseas with the Australian community. No prescribed qualifications are required to apply for a Churchill Fellowship and the subject of proposed projects is limitless, provided there is evidence of a benefit to Australia.
Application deadline: 28 April 2022
More information about this fellowship can be found here.

![[Closes 24 Nov 2107] Apply now to the OpenPlant Fund!](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1509564315902-TUO4I6QRWI9TT8UGSIAJ/OpenPlantTwitter_400x400+%281%29.jpg)

![[Closes 7 Mar 2017] OpenPlant Research Associate (Haseloff Lab)](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1486552818859-FH76MCA8SMFU93WB85RX/OpenPlantTwitter_400x400.jpg)