As the final round of OpenPlant funding comes to a close, we look back on the past eight years of work contributing to academic excellence in plant synthetic biology, open and responsible research and creating a community of like-minded scientists across Cambridge and Norwich.
In 2015, OpenPlant was established as one of six National Synthetic Biology Research Centres. A collaboration between the University of Cambridge, the John Innes Centre and the Earlham Institute, OpenPlant was founded with three aims:
to create a hub for interdisciplinary exchange between Cambridge and Norwich, between the fundamental and applied sciences, that will underpin advances in UK agriculture and bioproduction.
to establish systems for the open exchange of new plant tools and DNA components that will promote commercial innovation and international scientific exchange.
to explore the wider implications of the technology at local and global scales. This will bring together a wide range of engineers, scientists and policy developers to explore new technologies and possible models for sustainable agriculture, bioproduction and land use.
Over the years, these aims have been achieved in various ways, including over 230 academic publications, 90 invited lectures, 130 collaborations, 320 engagement activities and 8 spin-outs, amongst others. To round up our achievements we wanted to highlight a few of the areas in which we believe OpenPlant has excelled and added immense value, both to the academic community and beyond.
Development of New Tools and Technologies
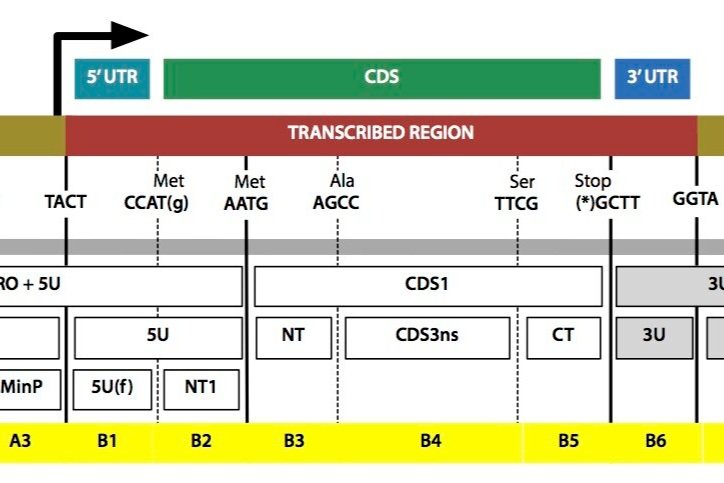
As set out in our original aims, the development of new tools for plant synthetic biology has been a core element of OpenPlant throughout the years. This aspect has focused not just on creating new tools, but also on ensuring that our research tools can be shared and freely used by academia and industry across the world. This “two-tier” model of innovation highlights the need for a shared base of basic resources that can be used to make innovation both more efficient and more equitable. OpenPlant has been instrumental in helping to develop this shared base of tools for the plant science community. Achievements include the development of multiple plant synthetic biology toolkits, including the OpenPlant toolkit, the Cyanogate toolkit and the Chlamydomonas reinhardtii MoClo toolkit, as well as the development of more efficient transformation systems in wheat. Additionally, the establishment of a common syntax for plant DNA parts and the development of the OpenMTA, have allowed open sharing and reuse of our materials.
Training and Career Development
Beyond our core research, OpenPlant has also worked hard to support our researchers with training and career development. Over the years, OpenPlant has employed 56 postdocs, students, research assistants and administrators, along with 23 group leaders. Of those, 23% have gone on to lead their own research groups, 16% have gone into industry, and 5% have gone into policy work. Workshops and initiatives to support our staff and students have included 14 technical training courses, 98 teams participating in the Biomaker Challenge, 73 teams funded by the OpenPlant Fund and two No-Code Training for Biology courses for researchers at Cambridge and Norwich.
Influence on Policy, Practice & the Public
In line with the OpenPlant ethos of open and responsible research and innovation, we have made a concerted effort to understand and translate our research in the wider context of society, into policy, education and public engagement. Projects such as DNA Dave and Global Garden Workshops, as well as our long-standing collaborations with the Science and Art (SAW) Trust have communicated work done by OpenPlant researchers to the general public, as well as inspiring young scientists to engage with art and science practice. The SynBio4Schools initiative has also brought plant synthetic biology into schools, introducing students and teachers to cutting edge research aligned with the UK national curriculum. OpenPlant researchers have also contributed to discussions on development of appropriate regulation for new genome editing technologies, for example through workshops and seminars for DEFRA, contributions to the Food Standards Agency advisory committee, horizon scanning workshop and public dialogue. OpenPlant working groups have also addressed the use of genetic resources in the age of the Nagoya Protocol, capacity building for the bioeconomy in Africa and the development of an Open Material Transfer Agreement.
Further Funding and Spin-outs
As a testament to the success of OpenPlant in academic excellence and collaboration building, OpenPlant research has attracted over £35.5 million in follow-on funding. This additional funding will allow the important work done by OpenPlant to continue, and has included funding for continuing collaborations between Cambridge and Norwich, including the Engineering Biology Transition Award granted to OpenPlant PIs Anne Osbourn, Nicola Patron, Jenny Molloy and Jim Haseloff. Moreover, OpenPlant is proud to have supported the establishment and growth of eight spin outs including Beneficial Bio, Colorifix, Persephone Bio, Leaf Expression Systems, Iceni Diagnostics, The Smarter Food Company, Tropic Biosciences and HotHouse BioEngineering.
Finally to say, the work of OpenPlant could not have been achieved without the commitment and dedication of our staff and students, and we would like to say thank you to everyone who has worked with, and engaged with, OpenPlant over the last eight years. OpenPlant has provided a core hub for interdisciplinary exchange between Cambridge and Norwich and we hope that collaborations between the OpenPlant partners continue to thrive and drive forward cutting edge and responsible research in future.

![[Closes 24 Nov 2107] Apply now to the OpenPlant Fund!](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1509564315902-TUO4I6QRWI9TT8UGSIAJ/OpenPlantTwitter_400x400+%281%29.jpg)

![[Closes 7 Mar 2017] OpenPlant Research Associate (Haseloff Lab)](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1486552818859-FH76MCA8SMFU93WB85RX/OpenPlantTwitter_400x400.jpg)