We are a pair of scientists at Medical Research Council Laboratory of Molecular Biology (MRC LMB), who are passionate about helping students learn about modern science.
Synthetic biology is particularly interesting to us as we both work at the forefront of this field and appreciate how biology has transformed into more of an engineering discipline, where we learn about life by building biological systems. The same principle, i.e., learning biology by doing it, is very efficient for studying complex concepts in schools. However, performing modern synthetic biology experiments in the classroom is an expensive activity, due to the reagents, media, bacteria and lab instruments needed, not to mention the paperwork burden of dealing with genetically modified organisms.
We believe that teaching modern science can be accessible, cheap and straightforward. We are not alone in this and there are significant developments that have been done by Amino Labs, Cell-free tech and Biobits, which pursue the same goal as us: to make cutting-edge science accessible and affordable. We chose to work with the cell-free transcription-translation system (TXTL) as it is cheap to make, there is no need for safety regulations and they are highly customizable: the only thing you need is a genetic construct.
Our aim is to teach students the principles of genetic control, the foundation of synthetic biology. The first thing that struck us was the ease with which children study electric circuits by directly connecting electrical parts in chains and experimenting with them. We wanted to reiterate this logic for biology. Luckily, the major principles of genetic regulation have already been established with electrical engineering in mind; the only puzzle piece missing: to connect them physically on a breadboard.
Figure A: Cell-free transcription-translation system (TXTL) using filter paper
The TXTL is meant to be a magic mixture that produces practically any genetic part (such as Green Fluorescent Protein or T7 RNA polymerase). As a material support where the reaction is contained, we chose a filter paper. The idea was to turn these pieces of paper into functional modules by expressing proteins in them. Therefore, connecting paper pieces later will let expressed proteins move from one paper piece to another with the water flow (Fig.A).
In the end, a protein expressed in one module can affect the reaction in the other. This experimental setup simplifies studying gene circuitry, as triggers and products of the circuit are physically separated and therefore theoretically it should be easier to deal with this kind of system as opposed to a black box mixture in the tube. Also, the possibilities are practically endless as this system is highly customizable and pieces could be connected in any way that should help children to experiment with material in an unconstrained manner.
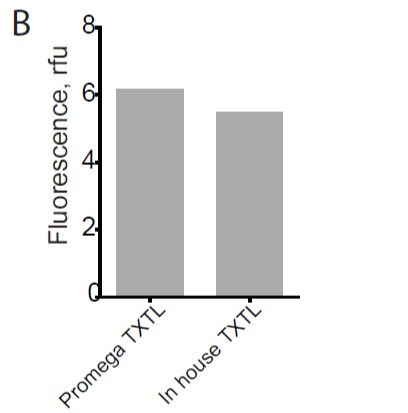
Figure B: comparison of activity between commercial mixture (Promega T7 high yield S30) and inhouse E.coli mixture.
The project started with the production of highly active TXTL E.coli mixtures. To help other laboratories that have access to only basic equipment, we used a cheap and easy protocol for preparing cell-extracts, so that our work is easily reproducible. We have prepared cell extracts either traditionally with a French press (Emulsiflex) and high-speed centrifuge, or using a cheaper and more streamlined approach by using ultrasound cell-lysis and a cooled table top centrifuge. Independent of the protocol we used for the preparation of E.coli lysate, activity was on par with the commercial mixture (Promega T7 high yield S30) (Fig.B).
Figure c: TXTL mixtures showing more active in solution than on paper.
The challenges began when we tried to run the TXTL reaction on paper: the cell-free mixtures are always active in the solution, their paper-based counterpart only gives a low signal which could only be visualized with expensive instrumentation and thus could not be used in any low-resource environments (Fig.C).
For now, we have found a viable alternative that is suitable for outreach: as opposed to lyophilizing TXTL on the paper, we freeze-dry TXTL in the tube. Surprisingly, the reaction mix was as active as the original one, and according to previous reports the reaction components retain their activity for weeks, and even months. Thus, we aim to use this ‘halfway’ TXTL product in the upcoming summer outreach. However, the battle is not over yet; we have now turned our attention to other support materials such as agarose, that does not interfere with TXTL, is cheap, could be freeze-dried and be cast in any form.
Follow the projects progress on twitter @zakir_tnimov