Marchantia as a model plant
Workpackage A: Simple Plant Chassis, Tools and Gene Delivery
OpenPlant is establishing Marchantia as a test bed for plant synthetic biology, exploiting its extraordinary experimental properties. This effort provides a prototype for other OpenPlant initiatives in higher plants. The aim of this Workpackage is to exploit the extraordinary experimental properties of Marchantia by producing systematic collections of (i) experimental protocols and (ii) shared DNA parts. This includes a comprehensive collection of promoters, selection markers, and fluorescent and biopigment reporter genes. In addition, (iii) we are producing and distributing Marchantia lines with integrated cell fate markers in order to track physiological and morphogenetic changes. We distribute information using laboratory-based web sites and online databases, and DNAs and plant material via the OpenMTA.
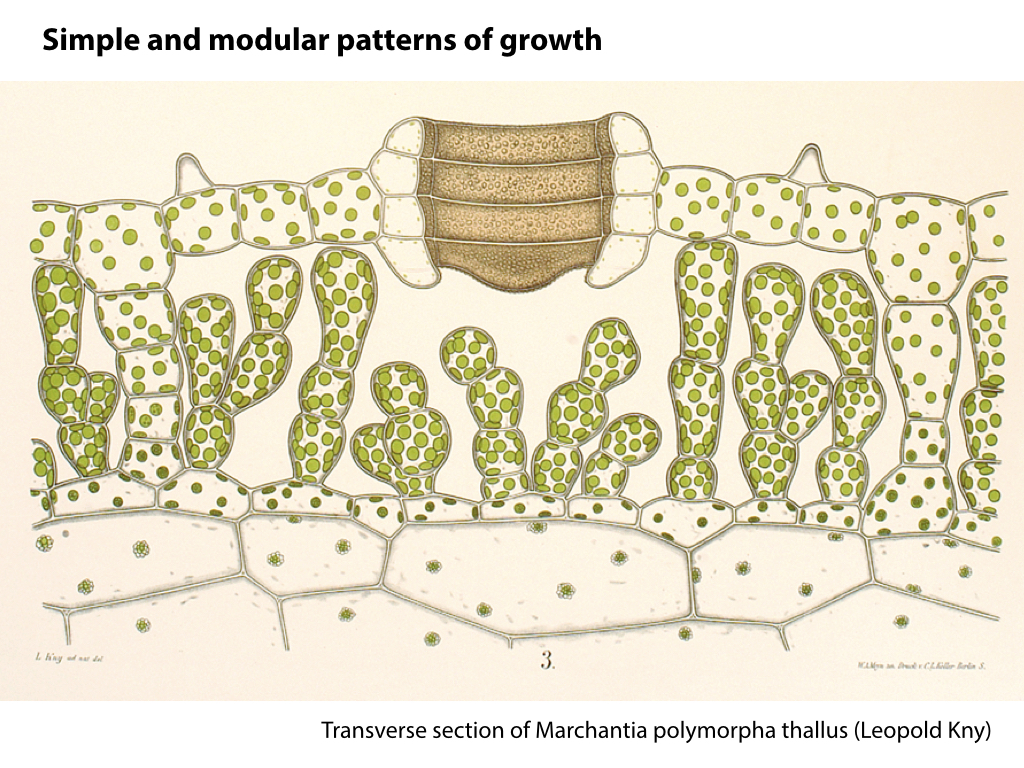
The liverworts (or Marchantiophyta) are descendants of the earliest terrestrial plants. The group is characterised by morphological simplicity, and this is matched by simple underlying genome structures. Many lower plants, including liverworts, demonstrate a striking tolerance of extreme stresses, a trait that would be valuable in a production system. Liverworts have been a largely neglected area of plant biology, but show great potential as new experimental systems after recent developments in transformation methods and genome characterisation.
Marchantia polymorpha is the best characterised liverwort plant. It is a common weed, and can grow quickly and resiliently. The relative simplicity of genetic networks in Marchantia, combined with the growing set of genetic manipulation, culture and microscopy techniques, are set to make this primitive plant a major new system for analysis and engineering.
Major benefits of working with Marchantia are:
The plant is a weed-like, small and easy to culture in soil, other media, and in axenic in vitro culture.
The thalloid liverwort is a descendant of earliest land plants, with the haploid gametophyte form dominant in its lifecycle. This allows the exploitation of haploid genetic systems, like microbes.
Sexual reproduction in male and female plants can be induced by far red light. The gamete bearing structures are easily identified and used for crosses. A single genetic cross results in the formation of millions of spores, which are stable for >1 year.
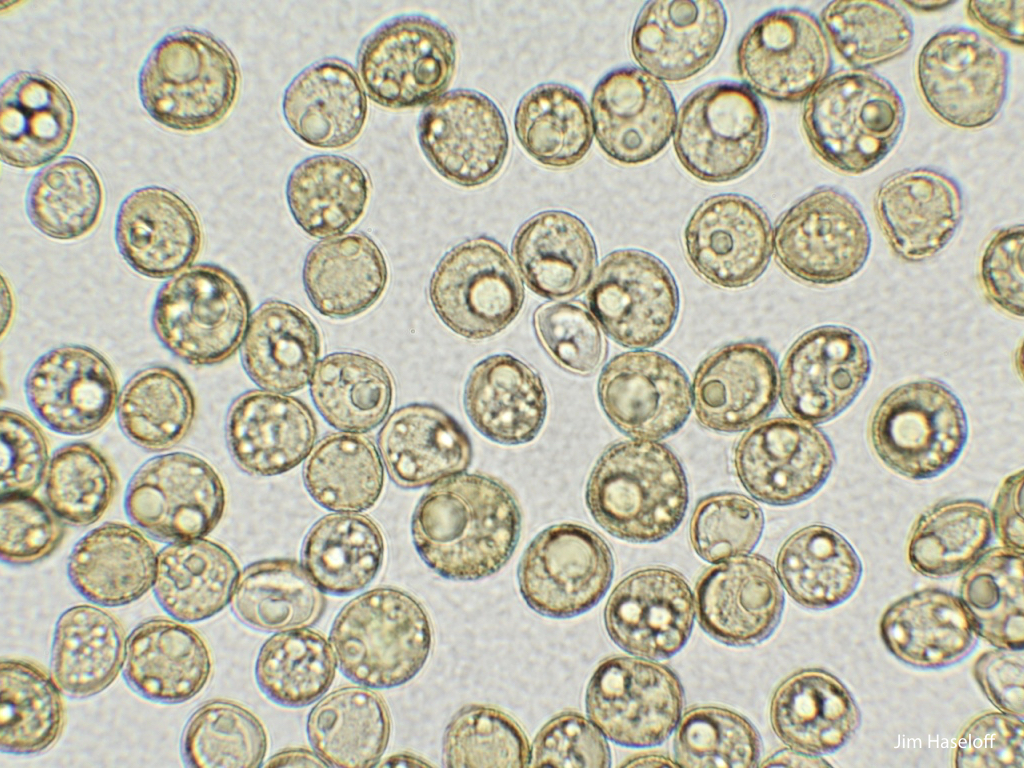
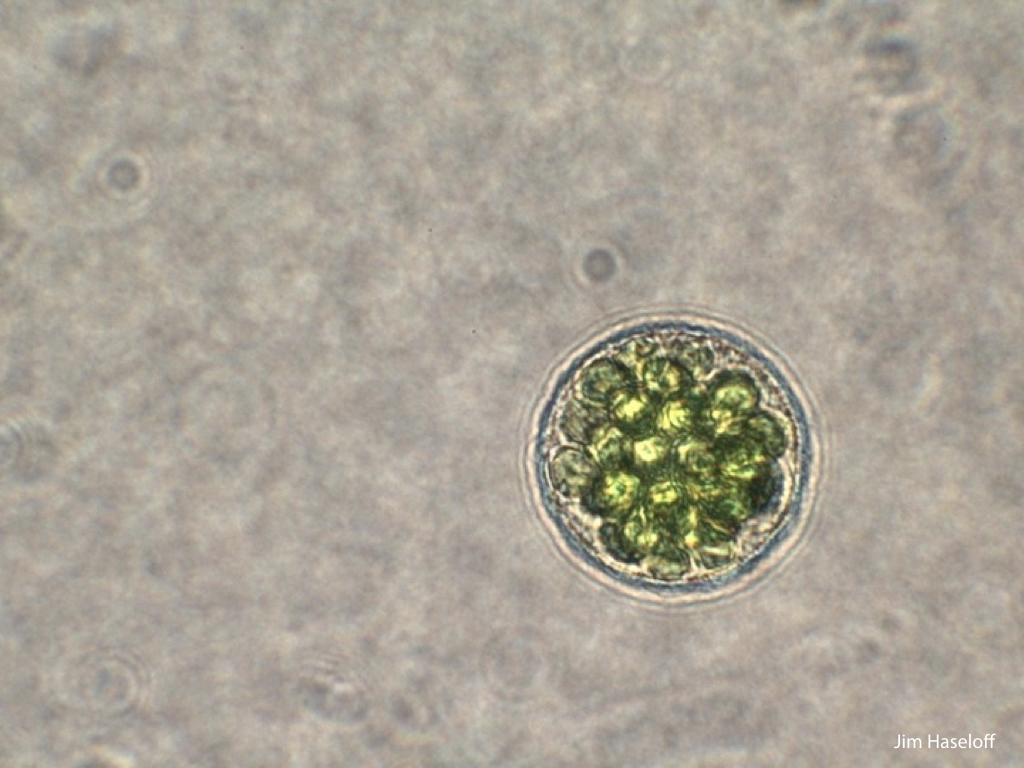
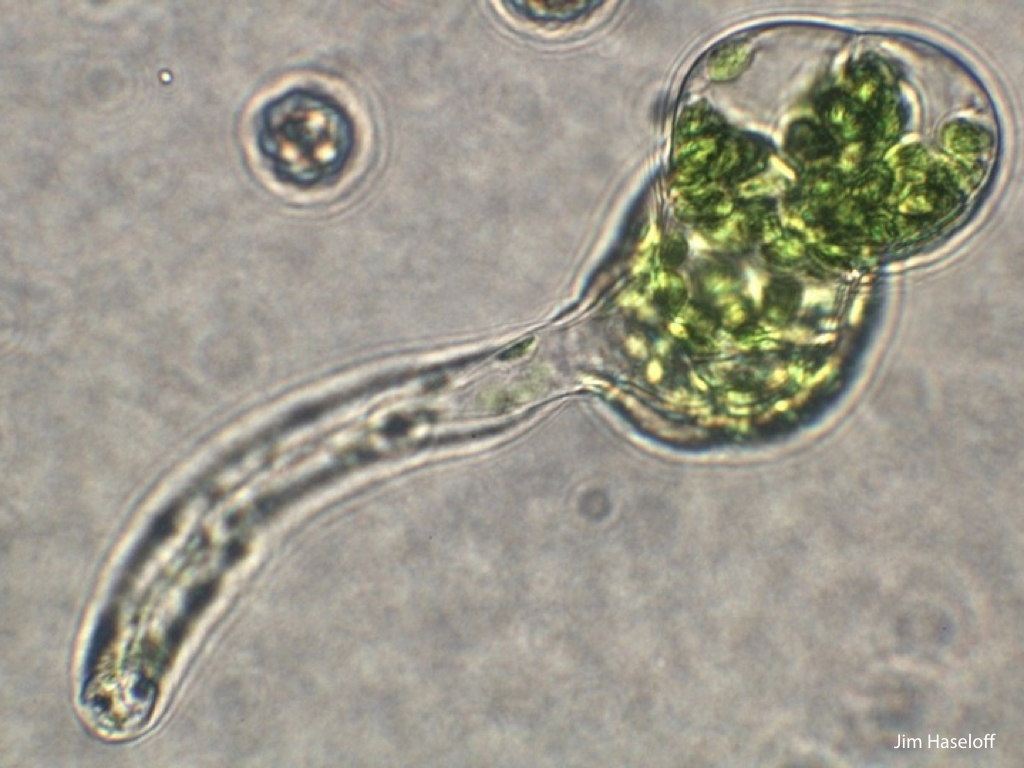
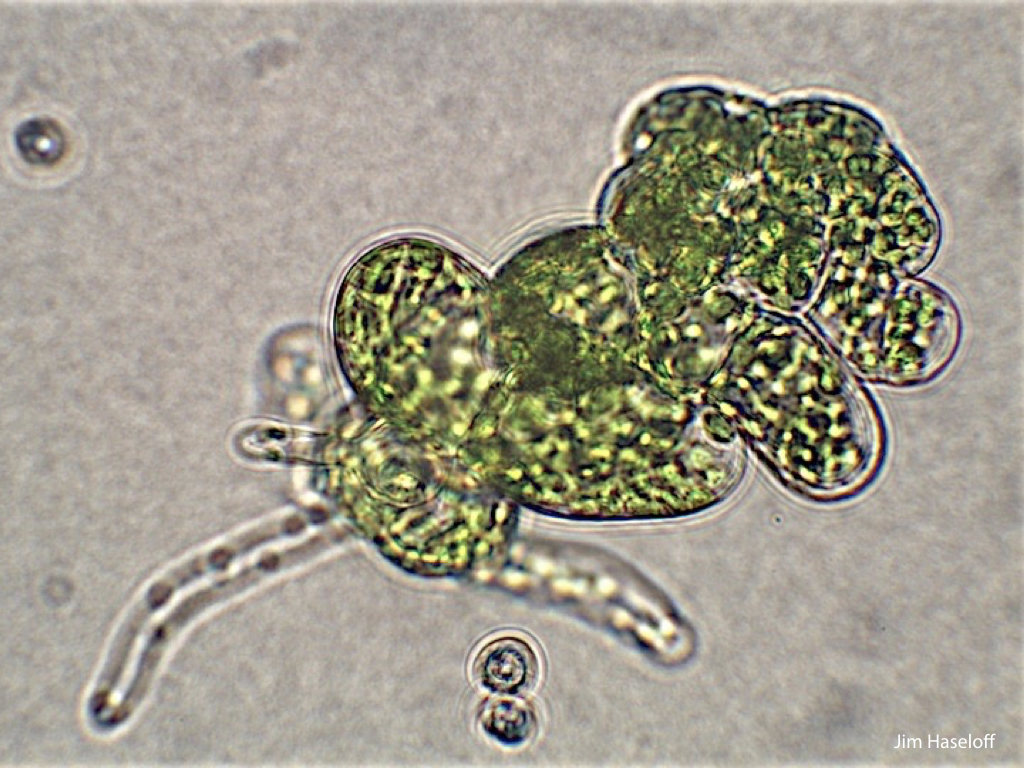
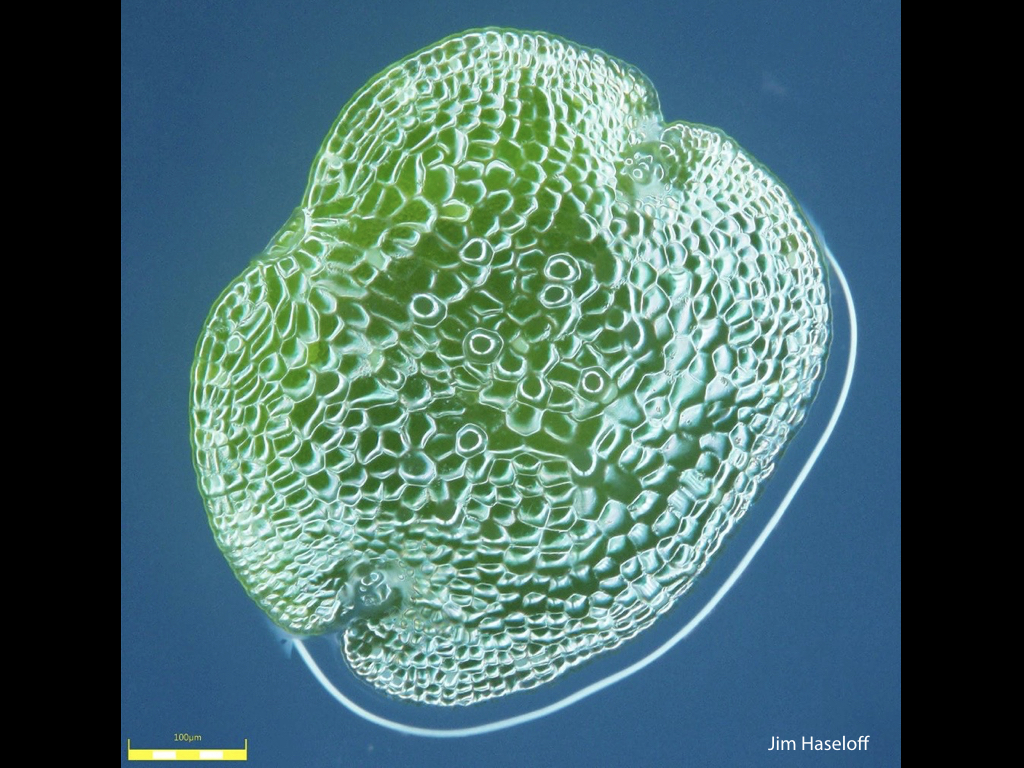
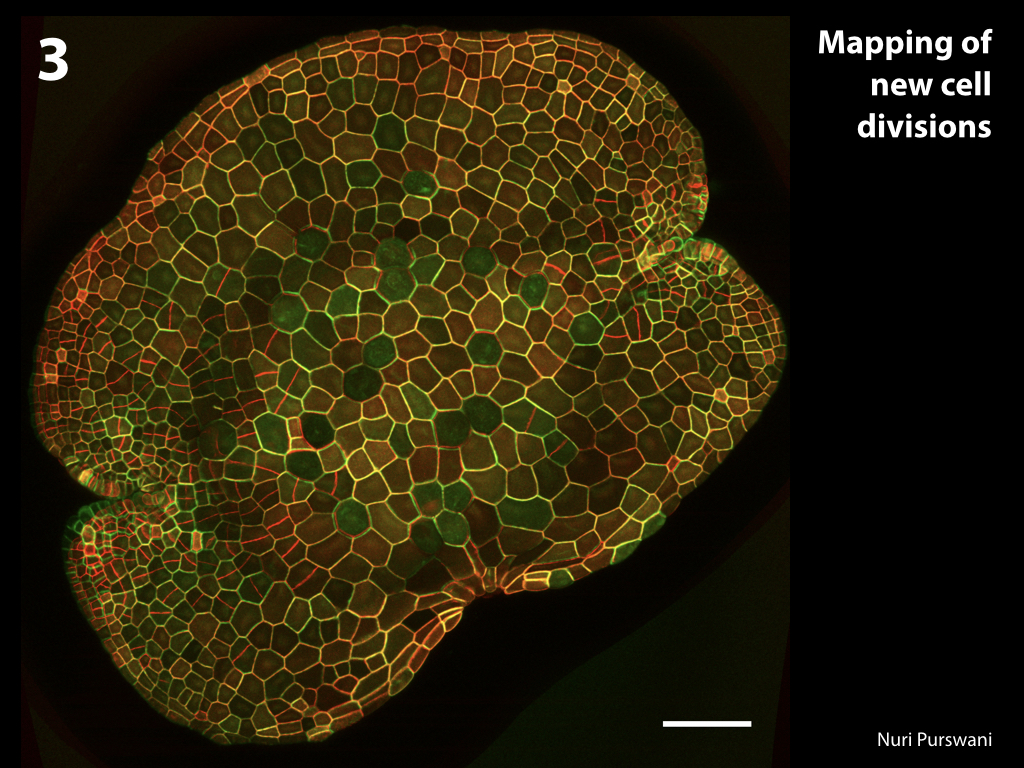
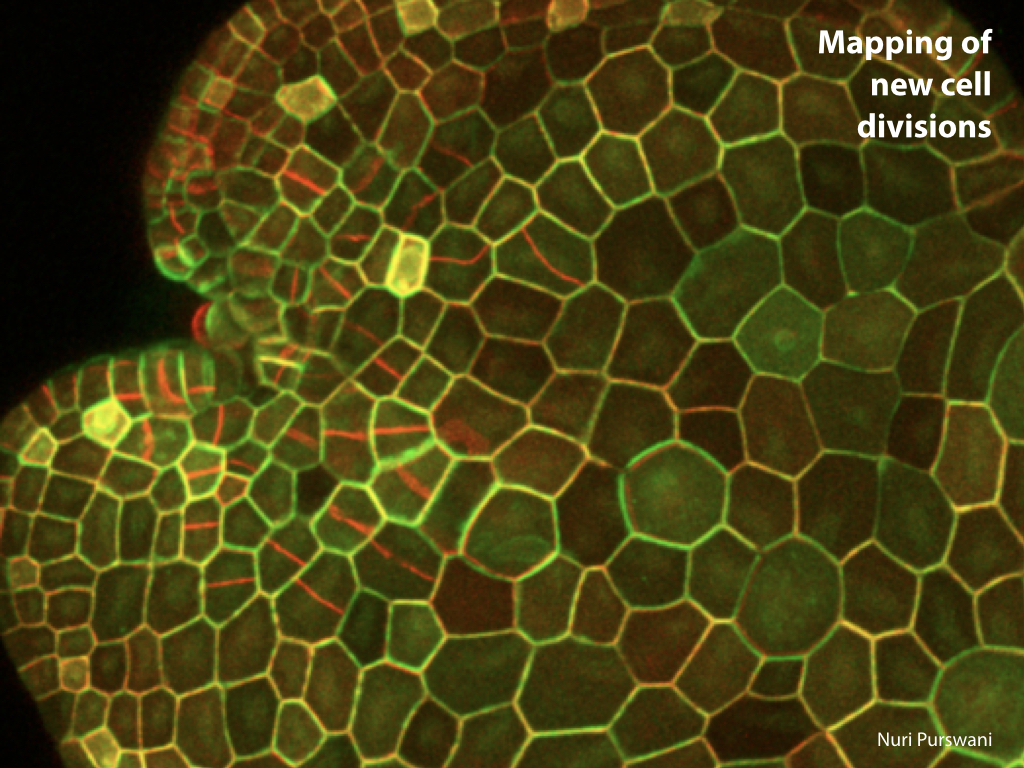
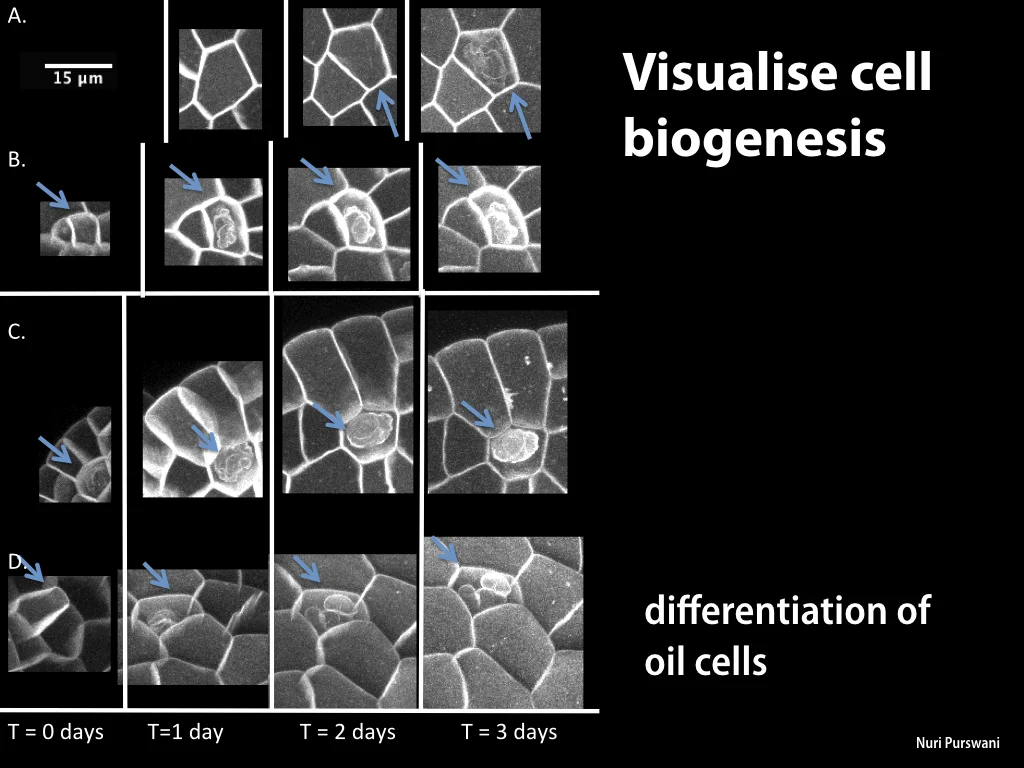
Spores will germinate and grow through the entire process of plant development in an entirely open fashion, accessible to modern quantitative microscopy techniques.
Marchantia plants spontaneously produce vegetative propagules, called gemmae, which allow simple clonal propagation of lines.
Gemma can be stored for 6 months as refrigerated stabs.
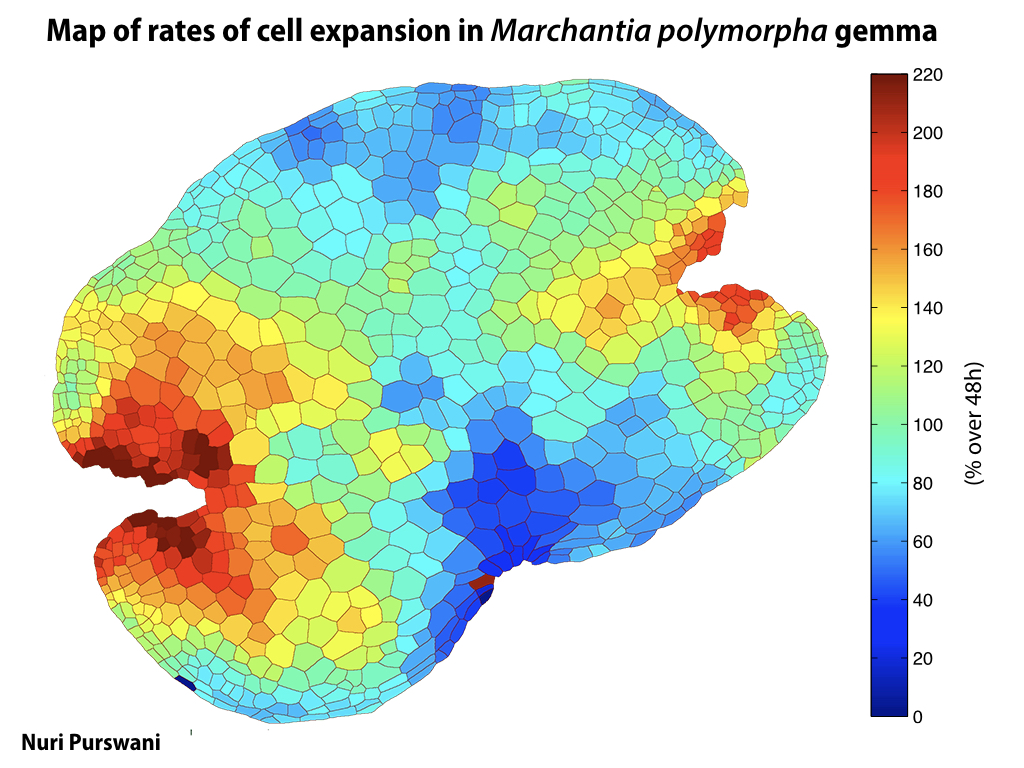
Gemma are ideal tissues for microscopic analysis of early development, with defined simple morphology and ability to rapidly progress through key developmental and physiological stages after germination.
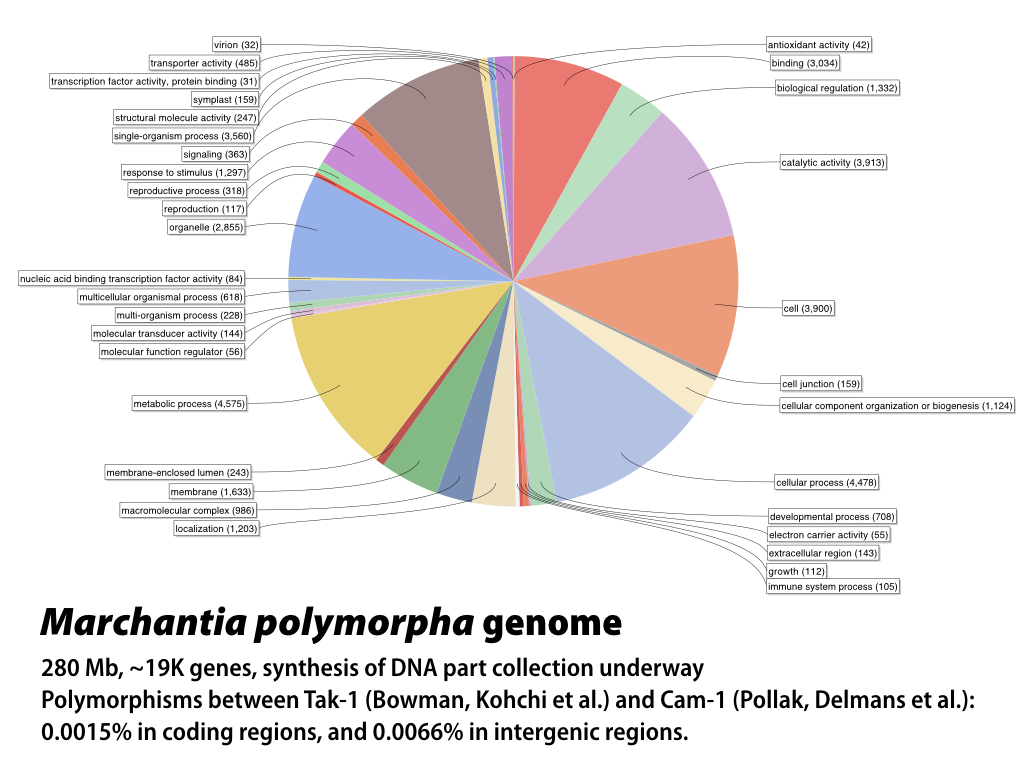
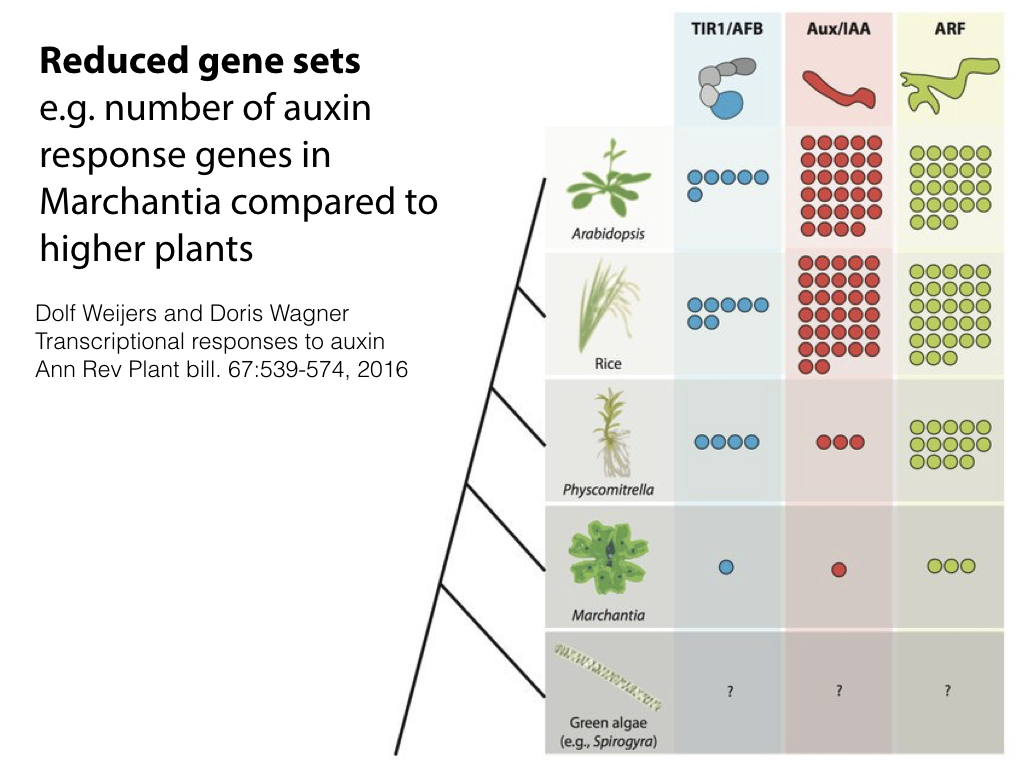
The 280 MB genome sequence is available in draft form at phytozome.net (Tak-1) and marpoDB.io (Cam-1). Compared to higher plants, Marchantia has a streamlined genome architecture, with reduced gene families.
Isolated Marchantia tissues easily regenerate to entire plants during tissue culture, allowing efficient plant nuclear and chloroplast transformation.
CRISPR-Cas9 based approaches for genome modification work efficiently and homologous recombination has been demonstrated in Marchantia.
Marchantia genome
In the Haseloff lab, Bernardo Pollak has generated draft genome and transcriptome sequences for the CAM-1 (male) and Cam-2 (female) isolates of Marchantia polymorpha. He and Mihails Delmans have annotated the genome, and constructed and published an online, gene-centric database (www.marpodb.io) that allows facile access to gene features from the M. polymorpha genome. The Oldroyd lab has generated genome sequence for M. paleacea (which, unlike M. polymorpha, forms symbiotic fungal associations). MarpoDB provides features that allow mining of the genome dataset for synthetic promoters, genes and terminators that can be exported directly as sequence files for synthesis of standardised Phytobrick DNA parts. The annotated genome sequence has provided a basis for comparison with the Tak-1 reference genome, which is best annotated and becoming the community standard (JGI resources) and (Marchantia.info). Polymorphisms between the Tak-1 and Cam-1/2 isolates are around 1/1000 bp within genes.
In addition, the annotated genome has allowed precise analysis of transcription dynamics in germinated Marchantia spores, where changes in mRNA production have been mapped over the crucial early stages of germination, cell enlargement, chloroplast differentiation, asymmetric cell division, rhizoid and thallus formation and cell differentiation. Sequence and transcriptome data have been contributed to the community-supported Marchantia genome effort.
The availability of the genome allows the extraction and testing of gene elements to generate a library of DNA parts for Marchantia.
Shared Resources
OpenPlant researchers are collecting protocols for work with Marchantia at protocols.io, for example:
Protoplast isolation: Marchantia protoplast preparation (adapted from Bopp and Vicktor 1988)
Spore transformation: This method produces random integration of DNA into the genome of Marchantia.
Media recipes: A range of media recipes for Marchantia growth
These can be found at https://www.protocols.io/groups/openplant-project
MarpoDB
MarpoDB provides features that allow mining of the genome dataset for synthetic promoters, genes and terminators that can be exported directly as sequence files for synthesis of standardised Phytobrick DNA parts.
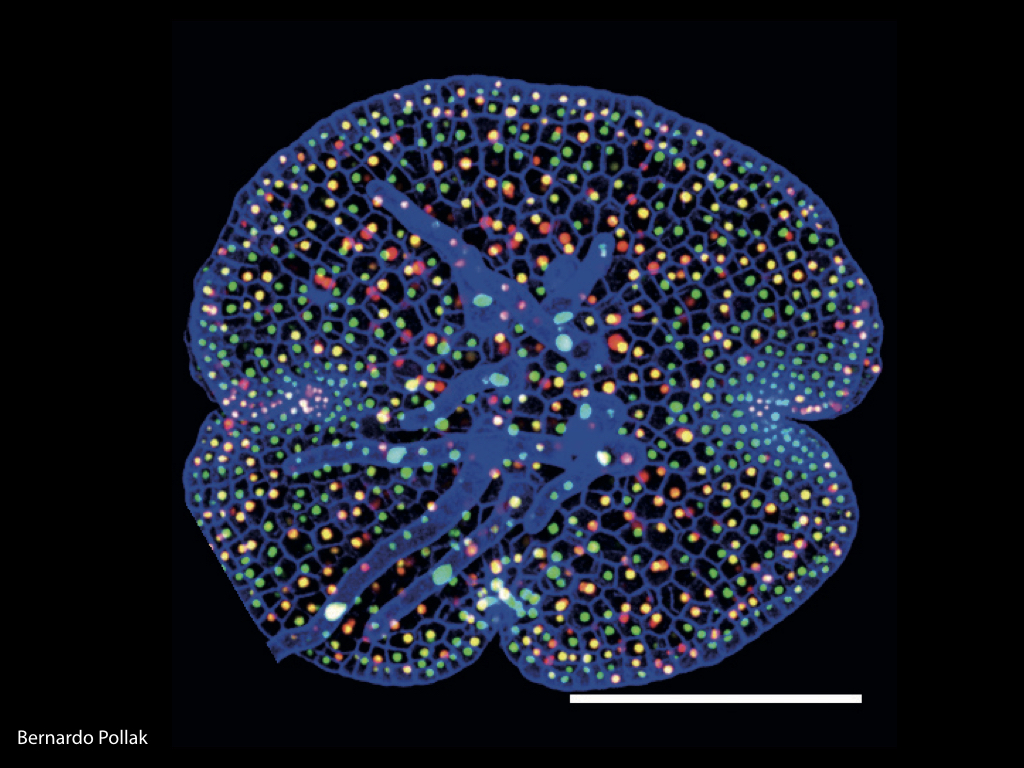
Candidate promoter sequences from genes homologous to meristematic growth in higher plants have been fused to fluorescent protein coding sequences and transformed into Marchantia. Ratiometric imaging techniques have validated the properties of the synthetic promoters and we are now in a position to scale and generate a comprehensive library of core promoters from all Marchantia regulatory genes. MarpoDB source-code is available on GitHub to promote development of computational tools for synthetic biology.
Distribution of Marchantia lines with integrated cell fate markers
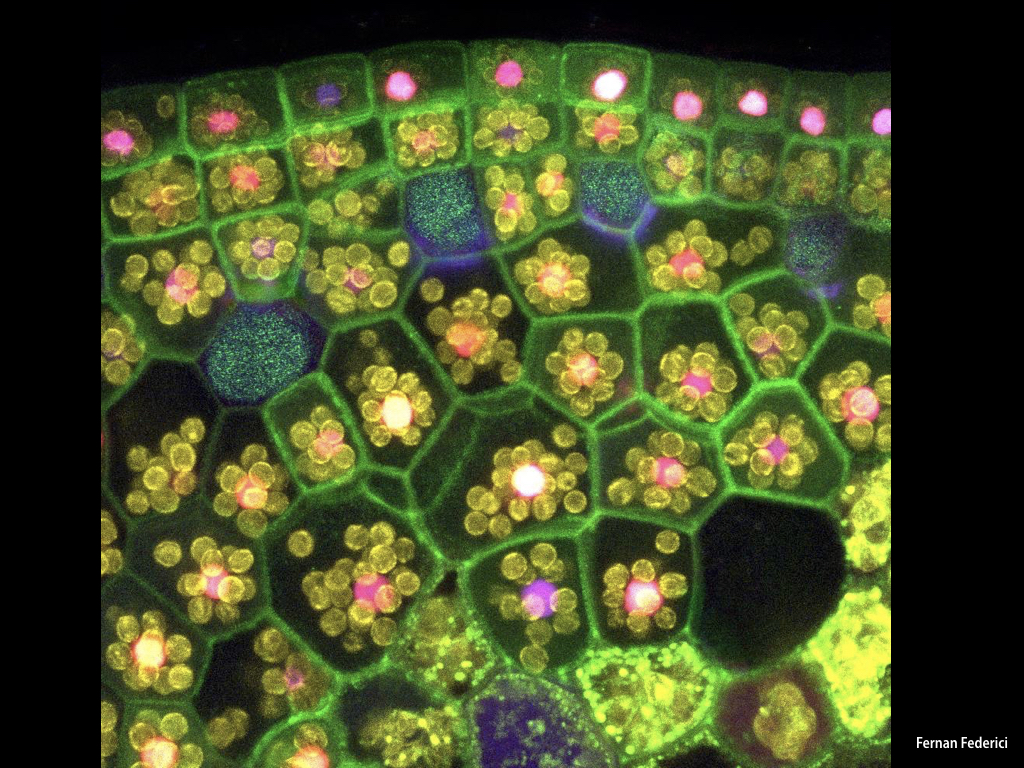
The Haseloff Lab has refactored spectral variants of fluorescent proteins for efficient expression in Marchantia, and developed a second and third generation GAL4 enhancer trap systems. The approach relies on the use of spectrally distinct fluorescent protein markers that allow autosegmentation of cell geometries and quantitative assignment of biological parameters on a cell by cell basis.
OpenPlant Toolkit for Marchantia
The first group of Marchantia resources have been recently published in ACS SynBio (Sauret-Gueto et al, 2020). It includes:
1) Techniques for simple and efficient propagation and maintenance of Marchantia lines throughout its life-cycle, with no requirement for specialized glasshouse facilities.
2) The type IIS OpenPlant DNA toolkit with Loop nuclear transformation vectors, Loop vectors for chloroplast transformation and Loop vectors for CRISPR genome editing. Also included is a collection of standardized DNA L0 parts for expression in Marchantia, including antibiotic resistance genes, signal peptides, fluorescent proteins for multispectral imaging and promoters for gene expression.
3) Techniques for high-throughput imaging of multiple Marchantia lines transformed with different fluorescent markers.
Marchantia is a morphologically simple plant with a dominant haploid phase and a fast life-cycle. It propagates sexually through spores and asexually through gemmae and can be easily maintained in sterile plates and boxes without requirement for glasshouse facilities. Type IIS DNA assembly methods, like Loop, allow for modular, hierarchical and standardized generation of DNA devices, assembling Level (L0) parts into transcriptional units (Level 1, L1) and L1 constructs into Level 2 (L2) devices. The OpenPlant toolkit contains the Loop vectors and a collection of Level 0 parts to use in Marchantia for nuclear and chloroplast transformation and CRISPR genome editing.
Online resources:
Publications:
Frangedakis F, Guzman-Chavez F, Rebmann M, Markel K, Yu Y, Perraki A, Tse SW, Liu Y, Rever J, Sauret-Gueto S, Goffinet B, Schneider H and Haseloff J (2020). Comparative analysis of early divergent land plants and construction of DNA tools for hyper-expression in Marchantia chloroplasts. BioRxiv 2020.11.27.401802 https://doi.org/10.1101/2020.11.27.401802
Sauret-Güeto S, Frangedakis E, Silvestri L, Rebmann M, Tomaselli M, Markel K, Delmans M, West A, Patron NJ, Haseloff J. (2020). Systematic tools for reprogramming plant gene expression in a simple model, Marchantia polymorpha. ACS Synth. Biol. 9, 4, 864–882 https://doi.org/10.1021/acssynbio.9b00511
Aguilar-Cruz A, Grimanelli D, Haseloff J, Arteaga-Vázquez MA. (2019) DNA methylation in Marchantia. New Phytologist, 223: 575-581
Pollak B, Cerda A, Delmans M, Álamos S, Moyano T, West A, Gutiérrez RA, Patron NJ, Federici F, Haseloff J (2018) Loop Assembly: a simple and open system for recursive fabrication of DNA circuits. New Phytol. 222(1):628-640. doi: 10.1111/nph.15625.
Kahl L, Molloy J, Patron N, Matthewman C, Haseloff J, Grewal D, Johnson R, Endy D. (2018) Opening options for material transfer. Nat Biotechnol. 36(10):923-927. doi: 10.1038/nbt.4263.
Boehm CR, Pollak B, Purswani N, Patron N, and Haseloff J (2017). Synthetic Botany. Cold Spring Harb Perspect Biol. 9(7). pii: a023887. doi: 10.1101/cshperspect.a023887.
Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, Adam C, Aki SS, Althoff F, Araki T, Arteaga-Vazquez MA, Balasubrmanian S, Barry K, Bauer D, Boehm CR, Briginshaw L, Caballero-Perez J, Catarino B, Chen F, Chiyoda S, Chovatia M, Davies KM, Delmans M, Demura T, Dierschke T, Dolan L, Dorantes-Acosta AE, Eklund DM, Florent SN, Flores-Sandoval E, Fujiyama A, Fukuzawa H, Galik B, Grimanelli D, Grimwood J, Grossniklaus U, Hamada T, Haseloff J, Hetherington AJ, Higo A, Hirakawa Y, Hundley HN, Ikeda Y, Inoue K, Inoue SI, Ishida S, Jia Q, Kakita M, Kanazawa T, Kawai Y, Kawashima T, Kennedy M, Kinose K, Kinoshita T, Kohara Y, Koide E, Komatsu K, Kopischke S, Kubo M, Kyozuka J, Lagercrantz U, Lin SS, Lindquist E, Lipzen AM, Lu CW, De Luna E, Martienssen RA, Minamino N, Mizutani M, Mizutani M, Mochizuki N, Monte I, Mosher R, Nagasaki H, Nakagami H, Naramoto S, Nishitani K, Ohtani M, Okamoto T, Okumura M, Phillips J, Pollak B, Reinders A, Rövekamp M, Sano R, Sawa S, Schmid MW, Shirakawa M, Solano R, Spunde A, Suetsugu N, Sugano S, Sugiyama A, Sun R, Suzuki Y, Takenaka M, Takezawa D, Tomogane H, Tsuzuki M, Ueda T, Umeda M, Ward JM, Watanabe Y, Yazaki K, Yokoyama R, Yoshitake Y, Yotsui I, Zachgo S, Schmutz J (2017). Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell 171(2):287-304.e15. doi: 10.1016/j.cell.2017.09.030
Boehm CR, Ueda M, Nishimura Y, Shikanai T, Haseloff J (2016). A Cyan Fluorescent Reporter Expressed from the Chloroplast Genome of Marchantia polymorpha. Plant Cell Physiol. 57(2):291-9. doi: 10.1093/pcp/pcv160.
Bowman JL, Araki T, Arteaga-Vazquez MA, Berger F, Dolan L, Haseloff J, Ishizaki K, Kyozuka J, Lin SS, Nagasaki H, Nakagami H, Nakajima K, Nakamura Y, OhashiIto K, Sawa S, Shimamura M, Solano R, Tsukaya H, Ueda T, Watanabe Y, Yamato KT, Zachgo S, Kohchi T (2016). The Naming of Names: Guidelines for Gene Nomenclature in Marchantia. Plant Cell Physiol. 57(2):257-61. doi: 10.1093/pcp/pcv193.