Science Practice’s interest in synthetic biology goes back to the Arsenic Biosensor Collaboration, and recently we were back in Jim Haseloff’s lab to learn about a new development called cell-free synthetic biology. This technology made headlines when it was used to create a low-cost, paper-based diagnostic test for the Zika virus. We’re interested in paper-based diagnostics (see SoilCards) and portable technology that could enable lab analysis in the field, like the MinION. Because of our work in this area were we asked to co-convene a workshop called “Programmable biology in the test tube”, which was organised by Jenny Molloy from the Strategic Research Initiative for Synthetic Biology at Cambridge.
This is a guest post by Dr Tempest van Schaik from Science Practice. Follow her @Dr_Tempest!
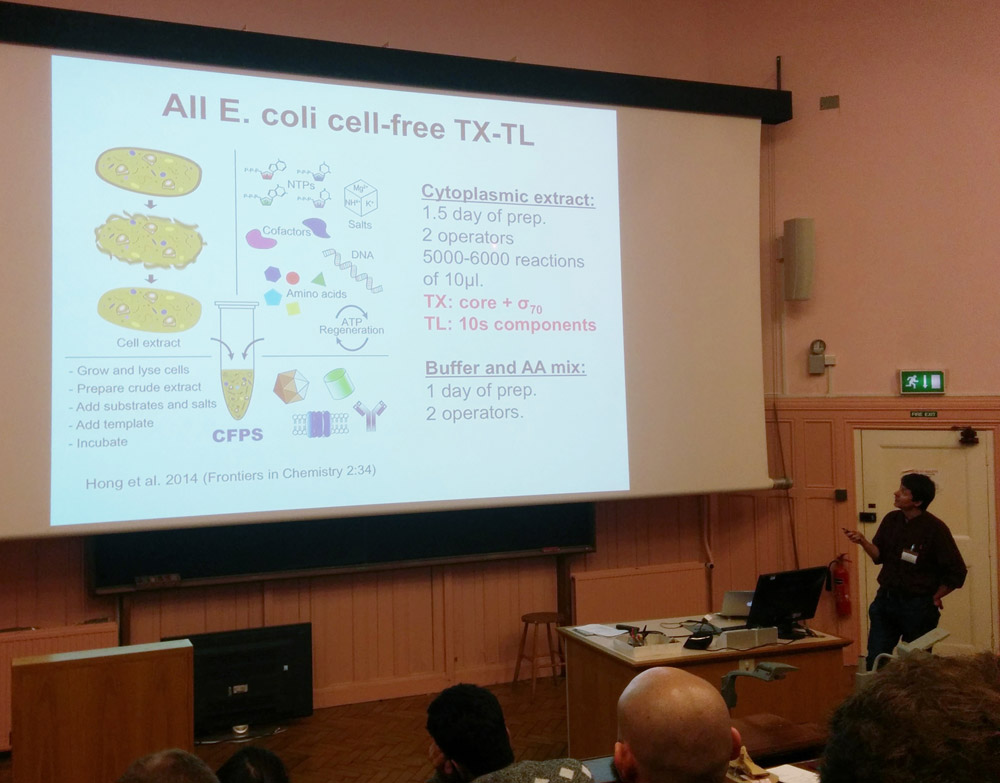
Traditionally, synthetic biology has involved genetically engineering bacteria to do our bidding: producing a useful protein (for example a protein that is fluorescent) from a blueprint (DNA in the form of a plasmid). Cell-free synthetic biology means extracting the machinery that bacteria like E. coli use to produce protein from the DNA blue-print (the machinery for transcription and translation), and leaving the cell behind. There are numerous technical reasons why this is preferable, but the real-world consequences that we’re most excited about are that devices that use synthetic biology sensors 1) don’t need to be kept refrigerated, which means they can be transported long distances to reach rural places, and 2) are not restricted in use in the same way as whole genetically modified organisms.
Our day in Cambridge University’s Department of Plant Sciences included talks and a workshop. True to the open, collaborative, multi-disciplinary spirit of the OpenPlant initiative, there was a diverse group of attendees from the bio-hacking community, and even those in software engineering and economics.
Vincent Noireaux presents his cell-free synthetic biology work at the workshop: Programmable Biology in the Test Tube
Things you see in a plant lab. Department of Plant Sciences, Cambridge University.
The workshop was led by Vincent Noireaux (University of Minnesota), and hosted by Jim Haseloffand Fernan Federici. We worked from Vincent’s paper, “The All E.coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology.”
FollowScience Practice @sciencepractice
Just add water...we're making GFP in a test tube! #cellfree#synbio @synbioSRI"
Each group of attendees was given a different cell-free gene-circuit to create. In the end we reviewed whether our gene-circuit was behaving correctly by using a microplate reader to look at the kinetics of expression of green fluorescent protein.
Kinetics of expression of green fluorescent protein produced by our gene-circuit
Groups gather round and discuss their results with Vincent.
It was easy to construct these gene-circuits on the lab bench, even for non-experts. We’re really excited to see a new generation of paper-based diagnostics that use cell-free synbio sensors. Thanks to Jim, Jenny, and Fernan for having us!

![[Closes 24 Nov 2107] Apply now to the OpenPlant Fund!](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1509564315902-TUO4I6QRWI9TT8UGSIAJ/OpenPlantTwitter_400x400+%281%29.jpg)

![[Closes 7 Mar 2017] OpenPlant Research Associate (Haseloff Lab)](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1486552818859-FH76MCA8SMFU93WB85RX/OpenPlantTwitter_400x400.jpg)